Data and metadata structures for time-resolved and multidimensional STEM
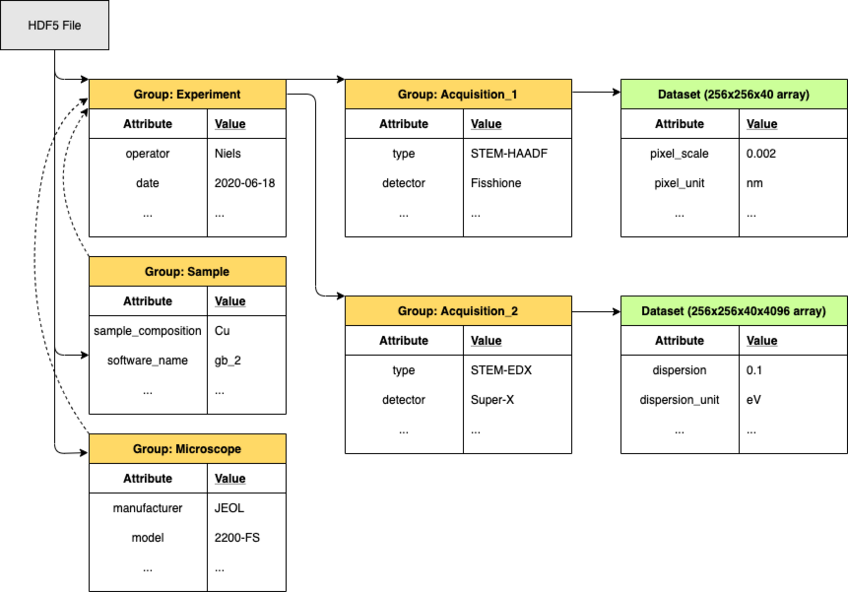
The formats must be self-descriptive, complete, yet flexible to be extended for novel experimental modalities. The data must interface easily with open source python tools that are widely used in the microscopy community, such as hyperspy, libertem, nion swift and pycroscopy. Finally, the aim is to integrate dataset metadata in the NOMAD materials database in order to make experimental datasets searchable, freely accessible and reusable. Translating tools will be written to easily convert proprietary formats into the open format. Currently, HDF5 based formats, such as NeXus, are envisioned due to their inherently nested structure, as shown in Figure 1. Developing a data format only makes sense when the community adopts it. Therefore, we are collaborating with the groups of Christoph Koch and Claudia Draxl at the Humboldt University of Berlin funded through the BiGmax network of the Max Planck Society to develop a joint standard for complex microscopy datasets and their integration into NOMAD.
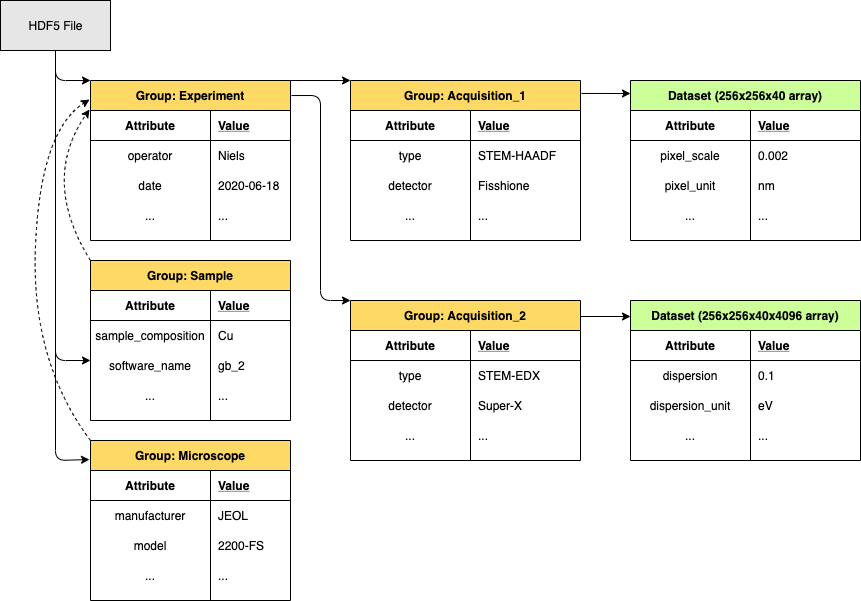
Figure 1: The internal structure of an HDF5 file is similar to a folder structure which allows for a logical and self-descriptive organisation of the data.